
Read our Privacy Notice if you are concerned with your privacy and how we handle personal information. If you plan to use these services during a course please contact us. If you have any feedback or encountered any issues please let us know via EMBL-EBI Support.
Nucleotide sequence analysis software#
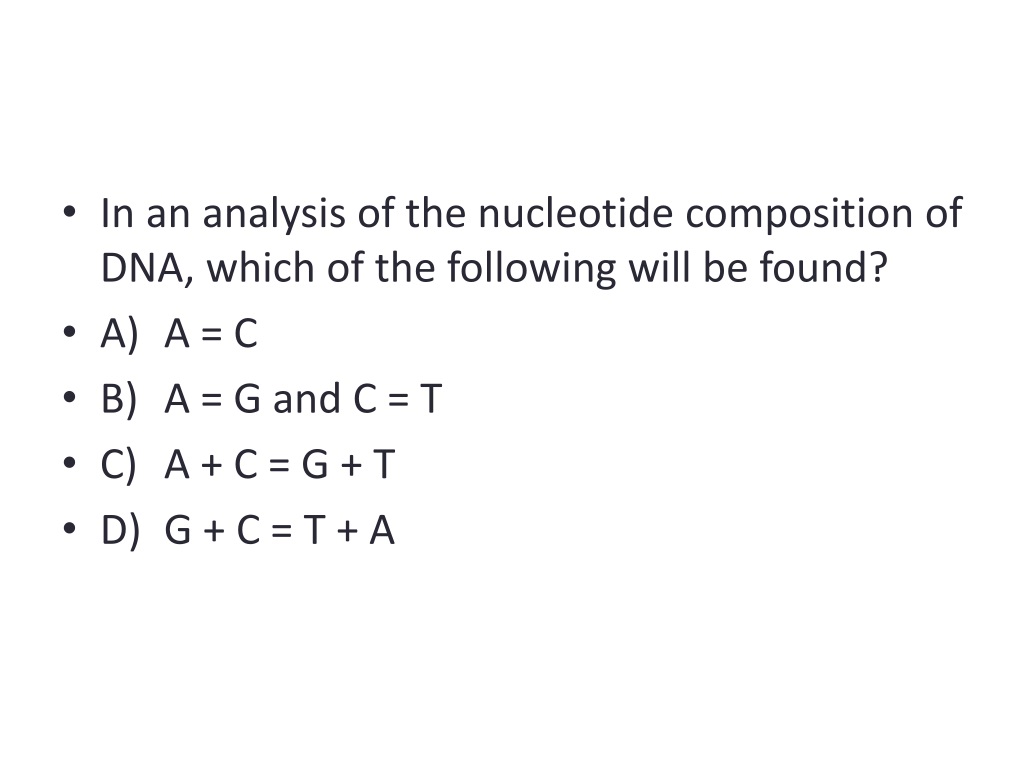
Please read the provided Help & Documentation and FAQs before seeking help from our support staff. The GenBank sequence format often has to be changed for use with sequence analysis software (Pearson and Lipman. The tools described on this page are provided using Search and sequence analysis tools services from EMBL-EBI in 2022 Launch Sixpack Protein Sequence Back-translationĮMBOSS Backtranseq back-translates protein sequences to nucleotide sequences.ĮMBOSS Backtranambig back-translates protein sequences to ambiguous nucleotide sequences. Nucleotide Sequence TranslationĮMBOSS Transeq translates nucleic acid sequences to the corresponding peptide sequences.ĮMBOSS Sixpack displays DNA sequences with 6-frame translation and ORFs. Read our Privacy Notice if you are concerned with your privacy and how we handle personal information.Sequence Translation is used to translate nucleic acid sequence to corresponding peptide sequences.īack-translation is used to predict the possible nucleic acid sequence that a specified peptide sequence has originated from.

Please read the provided Help & Documentation and FAQs before seeking help from our support staff. The EBI has a new phylogeny-aware multiple sequence alignment program which makes use of evolutionary information to help place insertions and deletions. Transform a Sequence Similarity Search result into a Multiple Sequence Alignment or reformat a Multiple Sequence Alignment using the MView program.Ĭonsistency-based MSA tool that attempts to mitigate the pitfalls of progressive alignment methods. Suitable for medium-large alignments.Īccurate MSA tool, especially good with proteins. MSA tool that uses Fast Fourier Transforms. In Sanger sequencing, the target DNA is copied. Very fast MSA tool that concentrates on local regions. DNA sequencing is the process of determining the sequence of nucleotides (As, Ts, Cs, and Gs) in a piece of DNA.
Suitable for medium-large alignments.ĮMBOSS Cons creates a consensus sequence from a protein or nucleotide multiple alignment. We have determined the nucleotide sequence of the Ri TL-DNA region from an Agrobacterium rhizogenes agropine-type plasmid using subcloned regions from the essentially identical Ri TL-DNAs from strains A4 and HRI. New MSA tool that uses seeded guide trees and HMM profile-profile techniques to generate alignments. The Addgene analyze sequence program is a tool for basic DNA sequence analysis that can detect common plasmid features in the sequence and create a map from. From the output, homology can be inferred and the evolutionary relationships between the sequences studied.īy contrast, Pairwise Sequence Alignment tools are used to identify regions of similarity that may indicate functional, structural and/or evolutionary relationships between two biological sequences. Multiple Sequence Alignment (MSA) is generally the alignment of three or more biological sequences (protein or nucleic acid) of similar length.


 0 kommentar(er)
0 kommentar(er)
